A broad-host-range synthetic sRNA platform was developed to effectively knockdown target genes in 15 bacterial species, including commensal, probiotic, pathogenic, and industrial bacteria, for medical and industrial applications.
Bacteria are intimately involved in the daily lives of humans. These microorganisms have not only been used in human history for food such as cheese, yogurt, and wine, but have also recently been used extensively as microbial cell factories to manufacture plastics, dietary supplements, and drugs. In addition to the benefits that bacteria provide to human life, pathogens such as Pneumonia and Staphylococcus, which cause various infectious diseases, are also ubiquitous. It is important to be able to metabolically control these beneficial industrial bacteria for high value-added chemical production and to manipulate harmful pathogens for pathogenicity suppression.
To address this issue, distinguished Professor Sang Yup Lee and his research team have developed a broad-host-range synthetic sRNA platform that can effectively knockdown target genes in 15 bacterial species including pathogenic, commensal, and industrial bacteria. The research team tested this platform tool for medical and metabolic engineering applications, demonstrating the mitigation of virulence-associated phenotypes in medical strains and the development of high-performance industrial strains. This knockdown tool will be useful to expedite the engineering of diverse bacteria that are of medical and industrial interest.
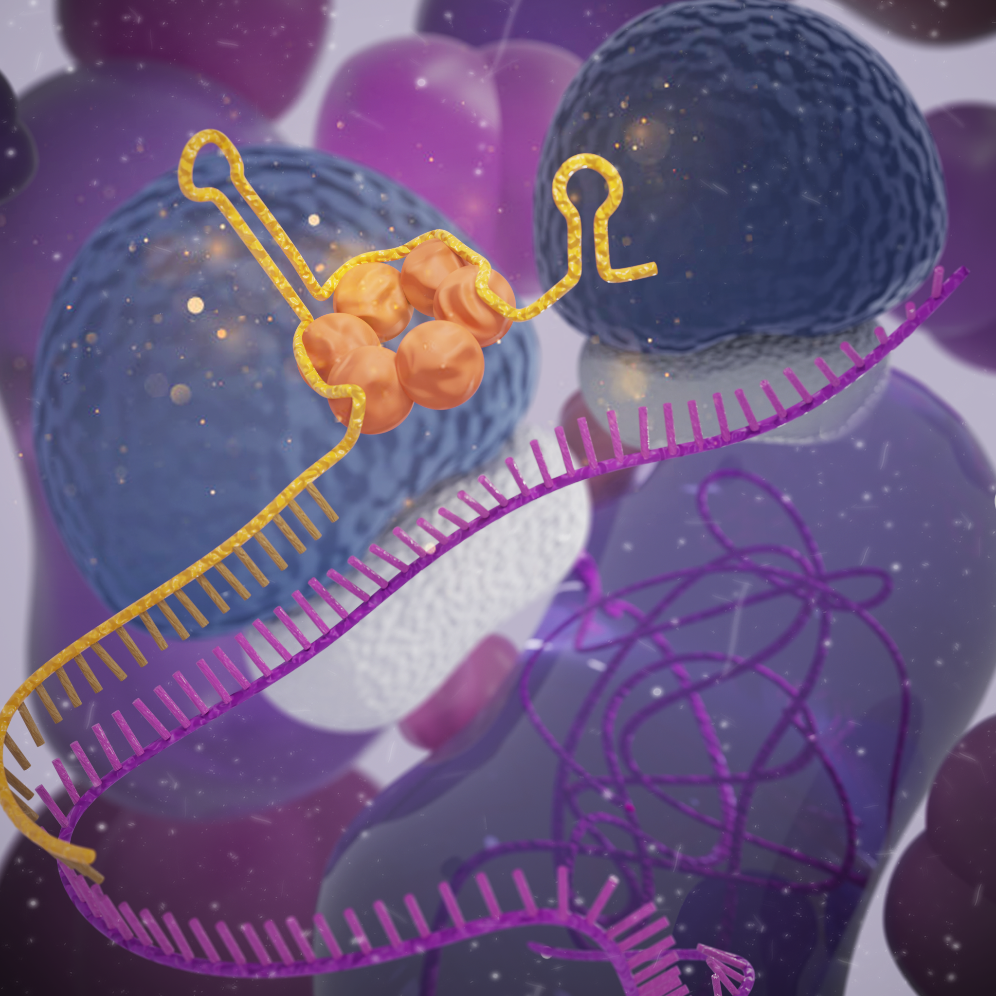
Previously, the team had developed a synthetic sRNA technique to knockdown genes in E. coli. Synthetic sRNA is a tool that can efficiently knockdown the expression of a target gene by binding to the mRNA of the gene, obstructing the ribosome from binding and consequently blocking the translation process. However, there was difficulty in implementing this system in Gram-positive bacteria as it was only effective in Gram-negative bacteria such as E. coli.
As a result, the research team developed a new sRNA platform tool that can effectively suppress target genes in various bacteria. After surveying sRNA systems in thousands of Gram-positive bacteria, an sRNA system derived from Bacillus subtilis was selected, as it showed the highest gene knockdown efficiency as a Broad-Host-Range sRNA (BHR-sRNA).
To address the problem of antibiotic-resistant pathogens, the BHR-sRNA was demonstrated to suppress the pathogenicity by suppressing the gene-producing virulence factor. By using BHR-sRNA, biofilm formation, one of the factors resulting in antibiotic resistance, was inhibited by 73% in Staphylococcus epidermidis, a pathogen that can cause hospital-acquired infections. Antibiotic resistance was also weakened by 58% in the pneumonia-causing bacteria Klebsiella pneumoniae. Additionally, BHR-sRNA was applied to industrial bacteria to produce high value-added chemicals including valerolactam, a raw material for polyamide polymers; methyl-anthranilate, a grape flavor additive; and indigoidine, a natural blue dye.
The BHR-sRNA developed through this study will help expedite the commercialization of bioprocesses to produce high value-added compounds and materials. It is also anticipated to help eradicate antibiotic-resistant pathogens in preparation for another upcoming pandemic. “In the past, we could only develop new tools for gene knockdown for each bacterium, but now we have developed a tool that can be used for various bacteria,” said Distinguished Professor Sang Yup Lee.
The results were published in the article titled “Targeted and high-throughput gene knockdown in diverse bacteria using synthetic sRNAs” online in Nature Communications on April 24, 2023.