KAIST
BREAKTHROUGHS
Research Webzine of the KAIST College of Engineering since 2014
Spring 2025 Vol. 24Novel creation of synthetic small regulatory RNAs enables the creation of high-throughput microbial biofactories
Novel creation of synthetic small regulatory RNAs enables the creation of high-throughput microbial biofactories
The development of microbial biofactories is receiving great interest worldwide for the eco-friendly and efficient production of chemicals, fuels, and materials. Synthetic small regulatory RNAs were developed for constructing microbial factories in a rapid and simple manner, and this synthetic sRNA-based strategy allowed the development of the most productive strains for two industrially important chemicals, tyrosine and cadaverine.
Article | Spring 2014
Today, people are concerned about global crises including global warming, fossil fuel depletion, and serious pollution. One of the ways to resolve the crises we are facing now is to develop sustainable technologies that can support us in daily lives and help us cope with the crises we face. One promising sustainable technology is microbial biofactories.
One of the major challenges in creating a biofactory is finding the best platform organism and gene(s) to engineer so as to maximize the biofactory’s efficiency to a level comparable to today’s refineries.
Recently, a Korean research team led by Distinguished Professor Sang Yup Lee at the Department of Chemical and Biomolecular Engineering of KAIST published their research results in in Nature Biotechnology (Feb, 2013) and Nature Protocols (Sep, 2013).
The advantages of utilizing synthetic small regulatory RNAs (sRNAs) are that they can be introduced easily into many different organisms in just one day since the blueprint (DNA) does not have to be modified. The efficiency of gene expression interruption can be modulated easily so that synthetic sRNAs can fine-control cellular gene expressions and can turn them on and off whenever an experimenter needs to do so. Synthetic sRNAs can be applied to control essential genes that cannot be deleted since the level of expression can be modulated. Finally, synthetic sRNAs allow any researcher to do large-scale experiments easily and rapidly without complicated processes or equipment. The paper, which was published as a journal cover paper in Nature Protocols, describes the detailed strategy and protocol for employing synthetic sRNAs for metabolic engineering.
Prof. Sang Yup Lee’s researchers applied the synthetic small regulatory RNA strategy to the production of tyrosine, which is used as a reliever of stress symptoms, a food supplements, and a precursor for many drugs. They examined a large number of genes related and unrelated to tyrosine production in multiple E. coli strains and succeeded to develop a biofactory with the highest yield ever. Through the strategy, they also developed another biofactory that produced cadaverine, an important platform chemical in the chemical industry, with high yield.
The new approach is expected to facilitate the development of biofactories that will resolve many inevitable problems stemming from industrialization. The continuous efforts in biofactory construction will enable the creation of small, sustainable, and environment-friendly biofactories instead of chemical factories causing environmental crises.
Reference: Dokyun Na, Seung Min Yoo, Hannah Chung, Hyegwon Park, Jin Hwan Park & Sang Yup Lee. Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nature Biotechnology 31, 170–174 (2013)
Most Popular

When and why do graph neural networks become powerful?
Read more
Smart Warnings: LLM-enabled personalized driver assistance
Read more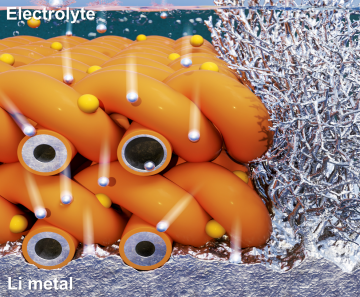
Extending the lifespan of next-generation lithium metal batteries with water
Read more
Professor Ki-Uk Kyung’s research team develops soft shape-morphing actuator capable of rapid 3D transformations
Read more
Oxynizer: Non-electric oxygen generator for developing countries
Read more
