KAIST
BREAKTHROUGHS
Research Webzine of the KAIST College of Engineering since 2014
Spring 2025 Vol. 24OncoSearch: A web tool that searches biomedical literature for meaningful pairs of cancers and genes
OncoSearch: A web tool that searches biomedical literature for meaningful pairs of cancers and genes
OncoSearch is a web tool that allows the user to query into biomedical literature for information on cancer-related genes and shows the results for further insights into oncogenesis, with an aim to catalyze and accelerate ongoing cancer research.
Article | Fall 2014
Automatic identification of gene-cancer relations from a very large volume of biomedical text is an important task for cancer research since changes in genes are known to be the main cause of oncogenesis, and a huge amount of information on such genes is archived in biomedical literature databases. To identify such relations, it is essential to understand, as much as possible, how a gene affects a cancer and to distinguish oncogenes (genes that cause cancers), tumor suppressor genes (genes that protect cells from cancers), and biomarkers (genes that indicate normal or cancerous states) since this will speed up the development of treatment and diagnosis methods for cancer.
Although genes may sometimes be explicitly claimed as oncogenes or tumor suppressor genes in the biomedical text, it is more often the case that information on gene-cancer relations is conveyed only implicitly with detailed descriptions about gene and cancer properties. Consider the example of the sentence below.
WWOX overexpression induced apoptosis and suppressed prostate cancer growth in vitro and in vivo [PMID:17704139].
While the gene WWOX is a well-known tumor suppressor, the sentence above does not contain an explicit reference to the gene as such. Instead, the sentence gives information that helps to classify the gene WWOX as a tumor suppressor of prostate cancer through the following inference: 1) WWOX expression level is increased, 2) prostate cancer regresses when WWOX expression increases, and 3) there is causality between the change in WWOX and the change in prostate cancer. By combining the three pieces of information above, one may classify the gene WWOX as a tumor suppressor gene. Although a single sentence with such implicit information may not provide enough evidence to confirm a particular gene’s class, collecting a large amount of such information in the literature would certainly help to substantiate such a conclusion.
Prof. Jong C. Park’s research team at KAIST developed OncoSearch, a web tool that allows the user to query into the biomedical literature for free-text information on cancer-related genes and provides the results for further insights into oncogenesis, or the process by which normal cells are transformed into cancer cells. In particular, OncoSearch can classify genes into either oncogenes, tumor suppressor genes, or biomarkers by taking into account implicit information as well as explicit information on their roles. The tool characterizes gene-cancer relations described in biomedical text with 1) how a gene changes, 2) how a cancer changes, and 3) the causality between the gene and the cancer, and the tool infers the respective roles of genes in cancers. Through this classification, the research team showed that the tool can correctly pick out oncogenes and tumor suppressor genes already registered as such in biology databases. The research team also showed that only small portions, 6.87% and 3.76%, respectively, of the oncogenes and tumor suppressor genes in one of the de facto standard gene databases, UniProtKB, are registered in the list of oncogenes and tumor suppressor genes published by Vogelstein and colleagues (Science, 2013). This indicates either 1) that the process of identifying new oncogenes or tumor suppressor genes is still at an early stage or 2) that the exact definitions of oncogene and tumor suppressor gene are highly dependent upon each biology database. OncoSearch is, thus, expected to catalyze much more research in oncology since the tool can collect and infer information about novel oncogenes, tumor suppressor genes, and biomarkers from the rapidly growing body of the literature that does not necessarily contain explicit expressions such as oncogene and tumor suppressor gene.
The picture shows part of the sentences that describe the oncogenic activity of CTNNB1 as retrieved by OncoSearch. OncoSearch classifies the gene CTNNB1 (also known as beta-catenin) as an oncogene, based on 109 automatically collected sentences from MEDLINE, a database that contains journal citations and abstracts for biomedical literature from around the world, maintained by the US National Library of Medicine. While mutation of the gene CTNNB1 is known to induce several types of cancers including colorectal cancer and ovarian cancer, the gene is not yet registered as an oncogene (or as a proto-oncogene) in one of the de facto standard gene databases, or UniProtKB. However, we see that, in 2013, the gene was registered as an oncogene in the Vogelstein list, while all the 109 sentences identified by OncoSearch are from 90 biomedical articles published before 2013, confirming the discovery of the role of the gene in oncogenesis.
For more information, check out
Most Popular

When and why do graph neural networks become powerful?
Read more
Smart Warnings: LLM-enabled personalized driver assistance
Read more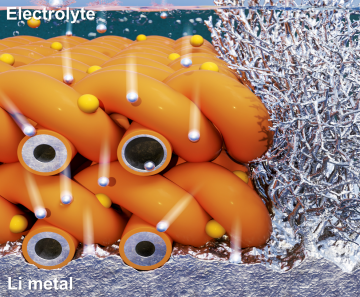
Extending the lifespan of next-generation lithium metal batteries with water
Read more
Professor Ki-Uk Kyung’s research team develops soft shape-morphing actuator capable of rapid 3D transformations
Read more
Oxynizer: Non-electric oxygen generator for developing countries
Read more
