KAIST
BREAKTHROUGHS
Research Webzine of the KAIST College of Engineering since 2014
Spring 2025 Vol. 24A new analysis algorithm reveals precise 3D spatial organization of the brain
A new analysis algorithm reveals precise 3D spatial organization of the brain
A new analysis technique developed for automated and calibrated neuron mapping into the standardized 3-D mouse brain reference atlas allows a precise analysis of the spatial organization of neural circuits and brings brain structure analysis to a new level.
Article | Fall 2020
A KAIST research team led by Professor Se-Bum Paik from the Department of Bio and Brain Engineering has developed a new analysis algorithm for the precise analysis of spatial organization of neural circuits, which brings brain structure analysis to a new level. This work was published in Cell Reports on May 26, 2020, under the title “Precise Mapping of Single Neurons by Calibrated 3D Reconstruction of Brain Slices Reveals Topographic Projection in Mouse Visual Cortex”.
AMaSiNe (Automated 3-D Mapping of Single Neurons), the developed algorithm, enables precise comparison of multiple brain data on a standard 3-D brain atlas, by calibrating misaligned and deformed slices using the minimum number of samples. This technique solves critical issues of existing algorithms that often require either massive whole-brain slicing data or the manual selection of atlas templates.
Brain tissue imaging is indispensable for neuroscience research. However, analysis of obtained imaging data mostly relies on manual processing, such as the visual matching of slice images to 2-D atlases, which cannot guarantee the accuracy and consistency of results. To address the issue, AMaSiNe, the new algorithm automatically calibrates the positions of slices in the standard brain atlas with only a few images, and reconstructs single neuron positions in a 3-D reference space, allowing direct comparison of data sets from different animals. It also accurately finds the alignment conditions from the distorted images and draws an accurate region of interest without any manual validation process. Furthermore, AMaSiNe uses versatile algorithms in fitting images of brain slices stained using various methods.
In collaboration with Professor Seung-Hee Lee’s group of the Department of Biological Science, the researchers exploited these benefits of AMaSiNe to investigate the topographic organization of neurons that project to the primary visual area, which could hardly be addressed without proper calibration and standardization of the brain slice samples by AMaSiNe. The results suggest that the precise correction of a slicing angle is essential for the investigation of important and complex brain structures.
The new algorithm is widely applicable and robustly used in studies of various brain regions and experimental conditions. For example, in a previous study led by the Yang Dan group at UC Berkeley, the current tool enabled accurate analysis of the neuronal subsets in the substantia nigra and their projections to the whole brain. This paper, which was co-authored by Professor Se-Bum Paik’s research group, was published in Science on Jan 24. Currently, the tool is of great interest to a wide range of scientists working in various fields of neuroscience and is being used by a number of research groups at MIT, Harvard, Caltech, UC San Diego, and KAIST.
The new algorithm allows a complicated spatial organization of neural circuits to be found in a 3-D brain, which would significantly impact analyses of brain slice images. More in-depth insights for understanding the function of brain circuits can be achieved by facilitating accurate and quantitative analysis of the spatial organization of neural circuits in the regions of the brain.
Most Popular

When and why do graph neural networks become powerful?
Read more
Smart Warnings: LLM-enabled personalized driver assistance
Read more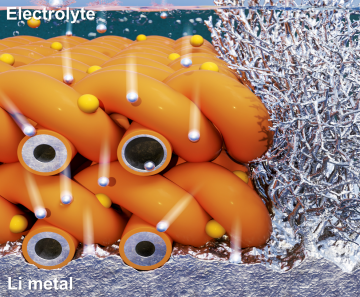
Extending the lifespan of next-generation lithium metal batteries with water
Read more
Professor Ki-Uk Kyung’s research team develops soft shape-morphing actuator capable of rapid 3D transformations
Read more
Oxynizer: Non-electric oxygen generator for developing countries
Read more



