KAIST
BREAKTHROUGHS
Research Webzine of the KAIST College of Engineering since 2014
Spring 2025 Vol. 24Deep learning predicts drug-drug and drug-food interactions
A deep learning-based computational framework called DeepDDI was developed, which predicts 86 drug-drug interaction (DDI) types with a mean accuracy of 92.4%. DeepDDI was used to suggest potential causal mechanisms for the reported adverse drug events of 12,721 drug pairs and also predicted alternative drug members for 62,707 drug pairs having negative health effects. Finally, DeepDDI was applied to 3,288,157 drug-food constituent pairs to predict drug-food interactions.
Article | Fall 2018
Drug interactions, including drug-drug interactions (DDIs) and drug-food constituent interactions (DFIs), can trigger unexpected pharmacological effects, including adverse drug events (ADEs) with often unknown causal mechanisms. However, current prediction methods do not provide sufficient details beyond the chance of DDI occurrence or require detailed drug information often unavailable for DDI prediction.
Assistant Professor Hyun Uk Kim and Distinguished Professor Sang Yup Lee both from the Department of Chemical and Biomolecular Engineering at Korea Advanced Institute of Science and Technology (KAIST) have developed a computational framework called DeepDDI that accurately predicts 86 DDI types for a given drug pair.
DeepDDI takes the structural information and names of two drugs in pair as inputs and predicts relevant DDI types for the input drug pair. DeepDDI uses a deep neural network to predict 86 DDI types with a mean accuracy of 92.4% using the DrugBank gold standard DDI dataset covering 192,284 DDIs contributed by 191,878 drug pairs. Importantly, DDI types predicted by DeepDDI are generated in the form of human-readable sentences as outputs, which describe changes in pharmacological effects and/or the risk of ADEs as a result of the interaction between two drugs in pair. For example, DeepDDI output sentences describing potential interactions between oxycodone (opioid pain medication) and atazanavir (antiretroviral medication) were generated as follows: “The metabolism of oxycodone can be decreased when combined with atazanavir”, and “The risk or severity of adverse effects can be increased when oxycodone is combined with atazanavir”. By doing this, DeepDDI can provide more specific information on drug interactions beyond the occurrence chance of DDIs or ADEs typically reported to date.
The research team used DeepDDI to predict DDI types of 2,329,561 drug pairs from all possible combinations of 2,159 approved drugs, and they found new DDI types of 487,632 drug pairs. They also showed that DeepDDI can be used to suggest which drug or food to avoid during medication in order to minimize the chance of ADEs or optimize the drug efficacy. To this end, DeepDDI was used to suggest potential causal mechanisms for the reported ADEs of 9,284 drug pairs and predict alternative drug candidates for 62,707 drug pairs having negative health effects to keep only the beneficial effects. Furthermore, DeepDDI was applied to 3,288,157 drug-food constituent pairs (2,159 approved drugs and 1,523 well-characterized food constituents) to predict DFIs. The effects of 256 food constituents on pharmacological effects of interacting drugs and bioactivities of 149 food constituents were also finally predicted. All these prediction results can be useful if an individual is taking medications for a specific (chronic) disease such as hypertension or diabetes mellitus type 2.
This research was published in Proc. Nat. Acad. Sci. (PNAS; 115, E4304-E4311, 2018) and was also highlighted in a number of news stories.
Most Popular

When and why do graph neural networks become powerful?
Read more
Smart Warnings: LLM-enabled personalized driver assistance
Read more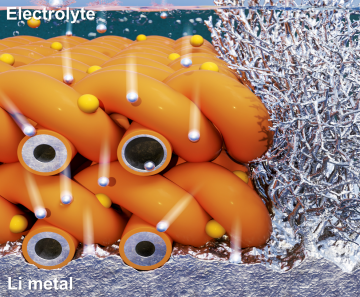
Extending the lifespan of next-generation lithium metal batteries with water
Read more
Professor Ki-Uk Kyung’s research team develops soft shape-morphing actuator capable of rapid 3D transformations
Read more
Oxynizer: Non-electric oxygen generator for developing countries
Read more
